U-Net++
UNet++ is a new versatile image segmentation architecture designed to improve image segmentation accuracy. UNet++ consists of U-Nets of varying depths, with decoders densely linked at the same resolution via revised skip routes.
By including Dense block and convolution layers between the encoder and decoder, it increases image segmentation accuracy
Architecture
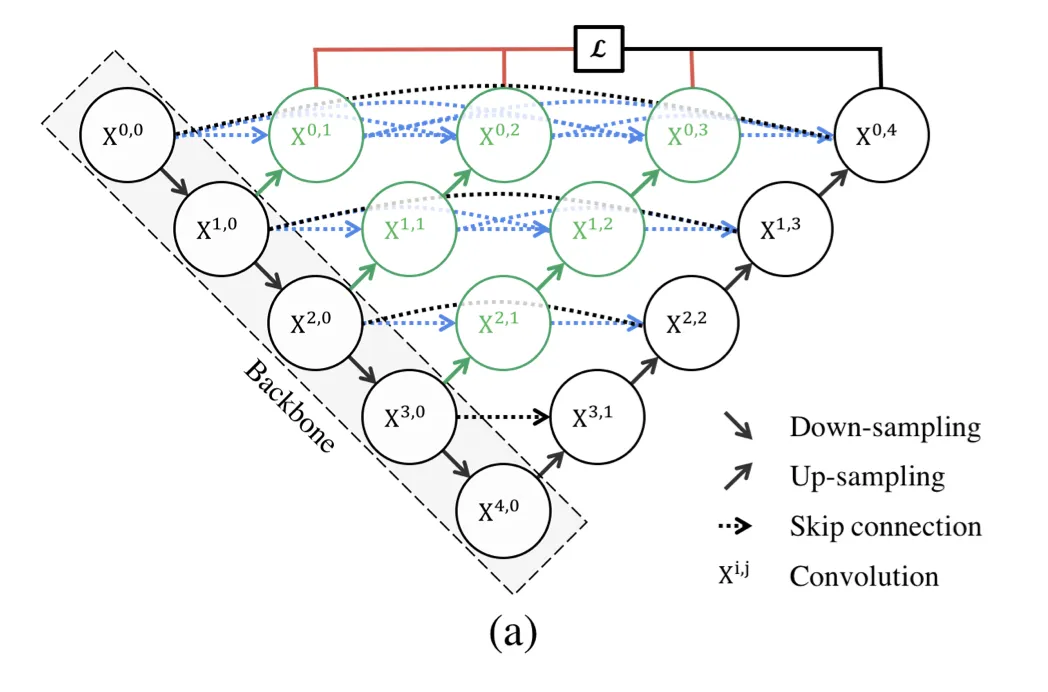
UNet++ starts with an encoder sub-network or backbone and then continues on to a decoder sub-network. The re-designed skip pathways (shown in green and blue) that connect the two sub-networks, as well as the use of deep supervision (shown red), separate UNet++ from UNet (black components).
Re-designed skip pathways
The main goal of introducing redesigned skip pathways is to bridge the semantic gap between the feature maps of the encoder and decoder subpaths. The hypothesis is that the optimizer would face an easier optimization problem when the received encoder feature maps and the corresponding decoder feature maps are semantically similar.
Deep supervision
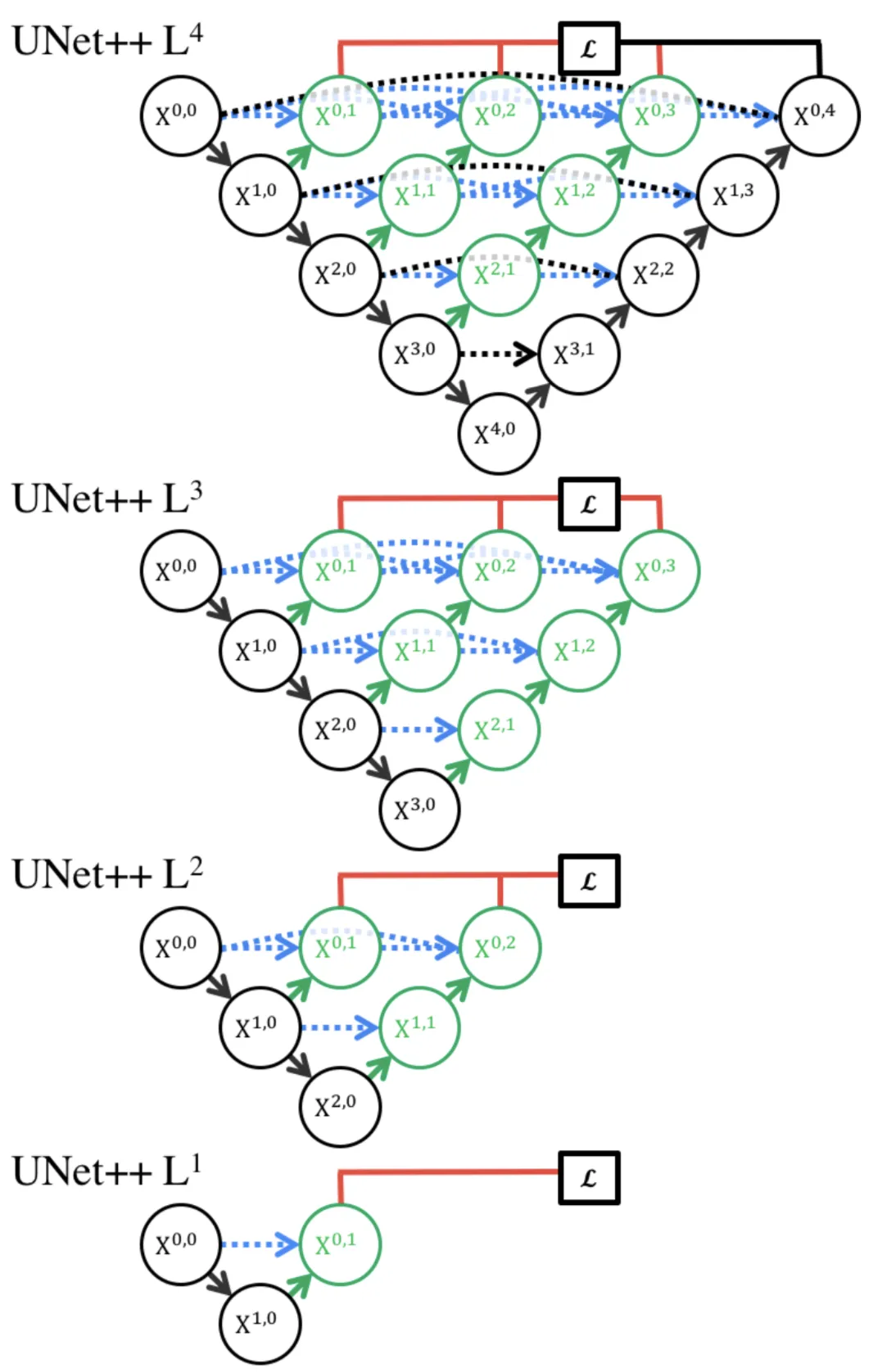
Deep supervision was introduced to the model so that it could decide the level of model pruning and speed gain.
This enabled the model to operate in two modes: 1) accurate mode wherein the outputs from all segmentation branches are averaged; 2) fast mode wherein the final segmentation map is selected from only one of the segmentation branches.
Parameters in model playground
Encoder network
In Model Playground, we can select feature extraction (encoding) network to use as either ResNet or EffiecientNet. The depth of the ResNet model/ EfficientNet sub type has to be specified as well.
Weights
After selecting the desired encoder network, the weights to be used for model initialization can be chosen.
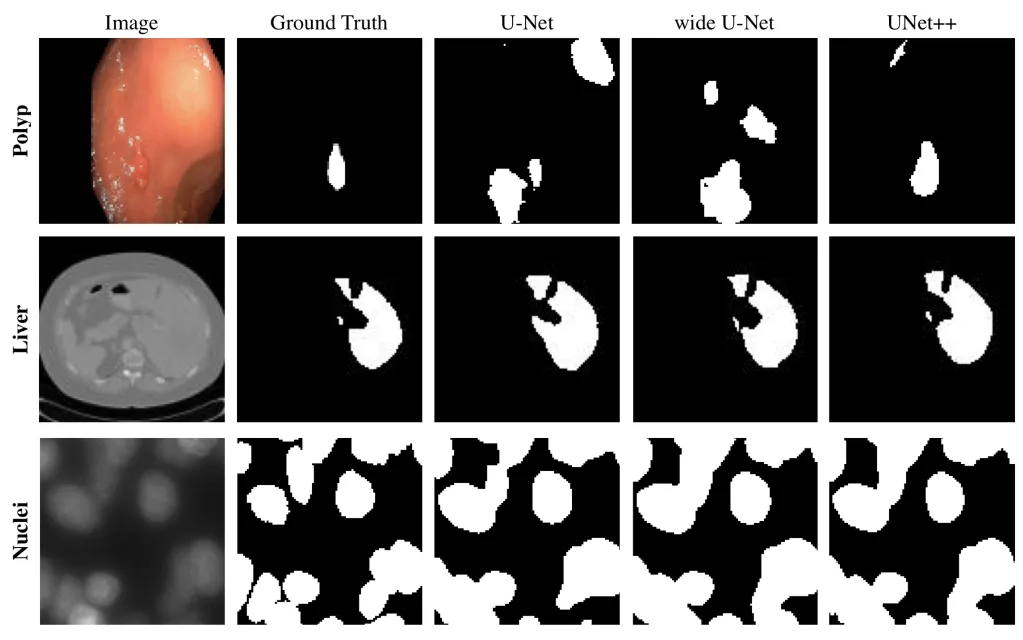
Qualitative comparison
Below the U-Net, wide U-Net, and UNet++ are compared showing segmentation results for polyp, liver, and cell nuclei datasets.
Further Resources
UNet++: A Nested U-Net Architecture for Medical Image Segmentation
